Abstract:
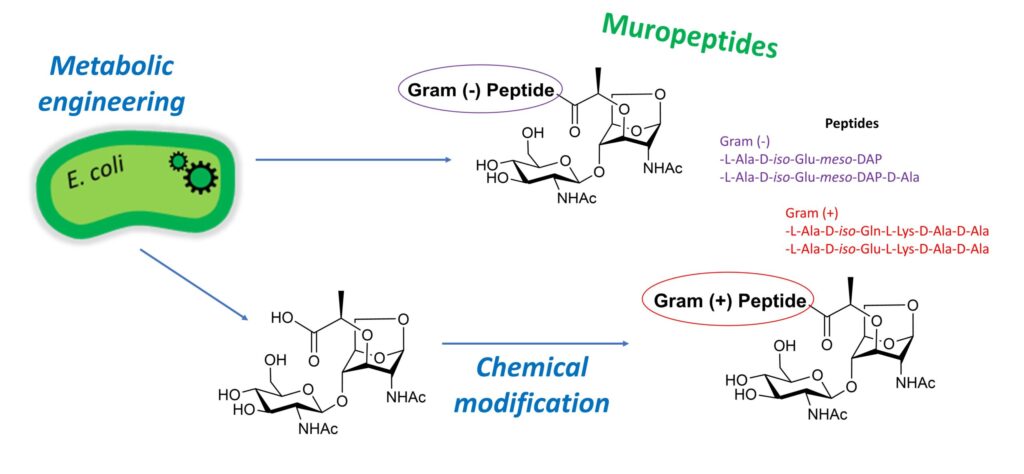
“Soluble fragments of peptidoglycan called muropeptides are released from the cell wall of bacteria as part of their metabolism or as a result of biological stresses. These compounds trigger immune responses in mammals and plants. In bacteria, they play a major role in the induction of antibiotic resistance. The development of efficient methods to produce muropeptides is therefore desirable both to address their mechanism of action and to design new antibacterial and immunostimulant agents. Here, we engineered the peptidoglycan recycling pathway of Escherichia coli to produce N-acetyl-β-D-glucosaminyl-(1→4)-1,6-anhydro-N-acetyl-β-D-muramic acid (GlcNAc-anhMurNAc), a common precursor of Gram-negative and Gram-positive muropeptides. Inactivation of the hexosaminidase nagZ gene allowed the efficient production of this key disaccharide, providing access to Gram-positive muropeptides through subsequent chemical peptide conjugation. E. coli strains deficient in both NagZ hexosaminidase and amidase activities further enabled the in vivo production of Gram-negative muropeptides containing meso-diaminopimelic acid, a rarely available amino acid.”
The article is available in open access overhere.